Stress response in biofilm forming Methicillin-resistant Staphylococcus aureus
DOI:
https://doi.org/10.62110/sciencein.jist.2025.v13.1178Keywords:
Methicillin-Resistant Staphylococcus aureus, MRSA, Biofilm, Stress response, Sigma factor B, Environmental stressAbstract
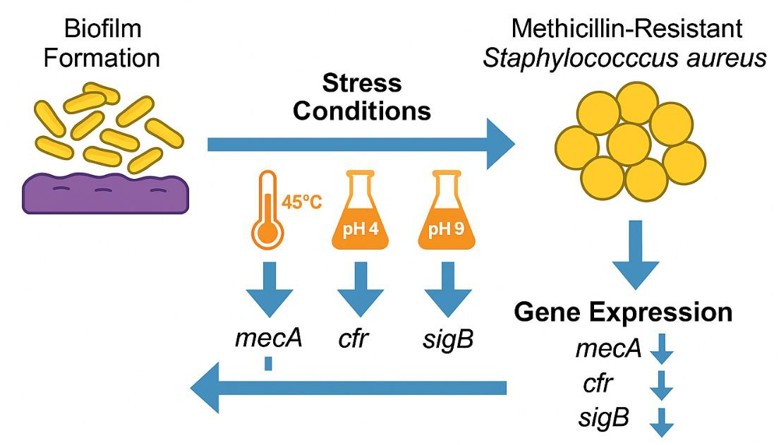
Staphylococcus aureus, especially methicillin-resistant strains (MRSA), represents a major global health concern due to its persistence, biofilm formation, and resistance mechanisms, with stress response pathways such as sigma B (sigB) playing a pivotal role in adaptation; however, their influence on biofilm dynamics and resistance gene expression in clinical isolates remains inadequately defined. This study aimed to assess the effects of thermal (45°C), acidic (pH 4), and alkaline (pH 9) stress on biofilm formation and the expression of key resistance/virulence genes (mecA, cfr, sigB) in MRSA isolates. A total of 20 MRSA strains, identified from 120 clinical samples (urine, blood, burns, and nasal swabs) collected at Al-Yarmuk Teaching Hospital between November 2024 and March 2025, were examined. Biofilm formation was evaluated using microtiter plate assays under both standard (37°C, pH 7) and stress conditions, while PCR confirmed the presence of mecA, sigB, and cfr, and qRT-PCR analyzed gene expression shifts using the ΔΔCt method with rpoB as a reference. Findings revealed that under normal conditions, 50% of isolates were strong biofilm formers; thermal stress decreased strong producers by 25% (p=0.025), acidic stress increased them by 80% (p=0.047), and alkaline stress reduced them by 35% (p=0.007). PCR positivity was high for mecA (90%), sigB (95%), and cfr (85%), while qRT-PCR indicated variable, isolate-specific gene regulation, with general downregulation of mecA and sigB under acidity and inconsistent expression responses under heat and alkalinity. Overall, biofilm formation in clinical MRSA is strongly influenced by environmental stresses, with acidity enhancing and heat/alkalinity suppressing it, while the widespread presence and variable expression of mecA, sigB, and cfr highlight the organism’s adaptive potential, complicating infection control and underscoring the need for continuous monitoring of these mechanisms to improve therapeutic strategies.
Downloads
Downloads
Published
Issue
Section
URN
License
Copyright (c) 2025 Manar T. Dakhel, Marwa H. M. Alkhafaji

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Rights and Permission







