Virtual screening of potential inhibitors against SARS-CoV-2 main proteases (Mpro) by dual docking with FRED and AutoDock Vina programs
Keywords:
Covid-19, SARS-COV-2, Mpro protease, virtual screening, FRED, AutoDock Vina, Molecular dockingAbstract
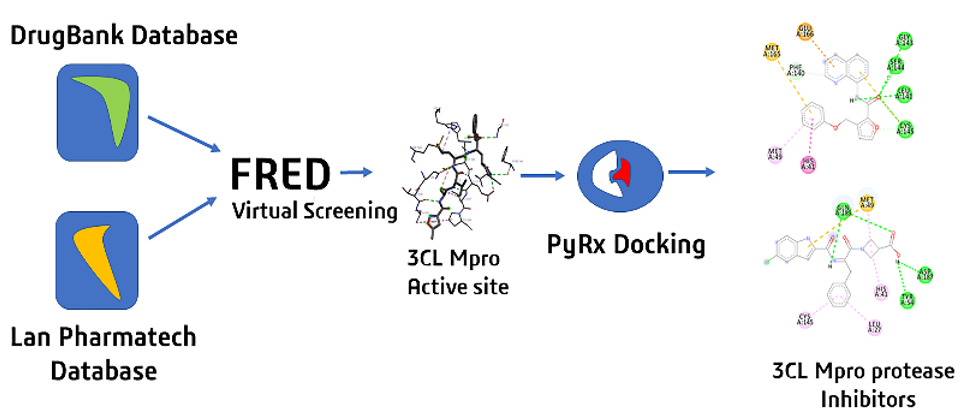
At this time, the COVID-19 (SAR-CoV-2) pandemic is still ongoing and considered the most serious global outbreak and we need drugs for treatment this virus. This study focused on searching existing drugs or compounds from PubChem database to provide a faster route for compounds to combat with COVID-19. Two databases from DrugBank (13,575 compounds) and Lan Pharmatech (72,350 compounds) were subjected to FRED docking program. Top five from the docking results of both databases had the binding energy ranging from -11.63 to -12.60 and -11.62 to -13.30 kcal/mol, respectively. These two sets of compounds were subjected to AutoDock Vina for fine tuning docking. The two best compounds from DrugBank interacted with catalytic Cys145-His41 dyad in 3CL Mpro protease, suggesting good binding pattern and energies. For Lan Pharmatech, two compounds demonstrated better protein-ligand interactions than the others. This dual docking protocol demonstrated fast and high efficiency in searching 3CL Mpro protease inhibitors.
Downloads
Published
Issue
Section
URN
License
Copyright (c) 2021 Prasan Tangyuenyongwatana, Wandee Gritsanapan

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
