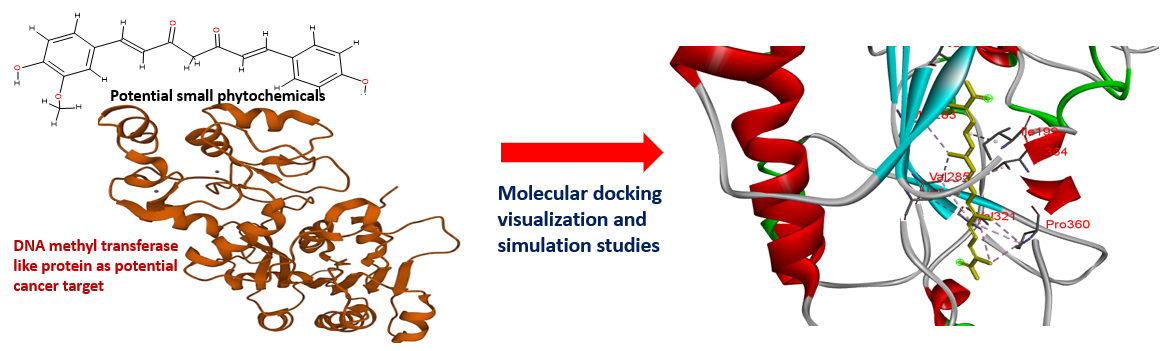
In silico identification of small natural inhibitors against DNA methyl transferase 3-like protein by integrative molecular docking and molecular dynamics approach
Keywords:
DNA methyl transferase 3 like protein, PASS, molecular docking, ADMETAbstract
Tiny phytochemicals found in different types of spices may play a significant role as anticancer drugs, particularly by focusing on a specific cancer receptor to modulate the cancer signaling cascade. We performed an in silico ADME and PASS analysis of twelve structurally different phytochemicals. Further, an integrative molecular docking and structural dynamics approach was carried out against a potential cancer target: DNA methyl transferase 3-like protein. Adopting an amalgamation of docking, simulation study, and MM-GBSA binding energy approach, we found carnosic acid (Ca) and crocetin (Cr) to be the best lead molecules. Hence, Ca and Cr may be proposed as strong potential anti-cancerous compounds against DNA methyl transferase3-like proteins.
URN:NBN:sciencein.cbl.2023.v10.537



