Establishment of in silico prediction methods for potential bitter molecules using the human T2R14 homology-model structure
Keywords:
Human T2R14, Bitter molecules, Docking simulation, Molecular dynamics, Machine learningAbstract
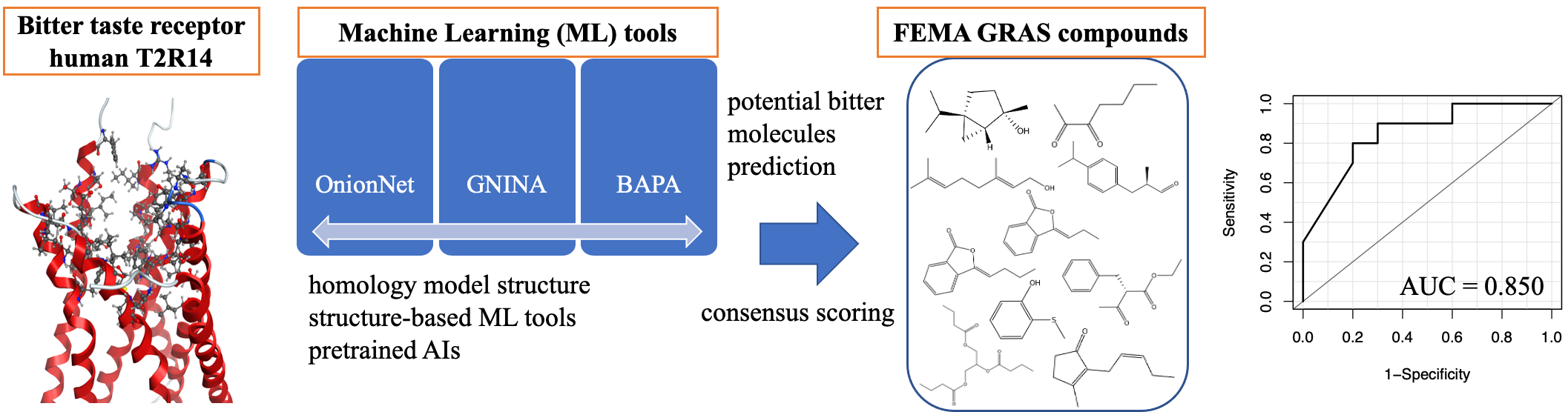
Bitterness is sensed by human taste receptors (hT2Rs) consisting of G protein-coupled receptors (GPCRs). The construction of an in silico evaluation system for bitter molecules using human T2R structure information will enable the identification of new bitter molecules and bitter blockers, which will contribute to food and drug development. Since the crystal structures of the hT2Rs have not been elucidated, we attempted to construct in silico discrimination methods for potential bitter molecules using the hT2R14 model structure in the GPCRdb that was constructed by the homology modelling method. Although the hT2R14 model structure was constructed using characteristics of existing bitter molecules, it was not previously clear whether it could be used for the prediction of new bitter molecules and bitter blockers. In this study, we established novel methods of predicting potential bitter molecule interactions with hT2R14 using datasets of compounds from ChemBridge and FEMA GRAS libraries. We used docking simulation tools, molecular dynamics simulation tools, structure-based machine learning (ML) tools, and sequence-based ML tools to establish potential bitter molecule prediction systems for hT2R14. Finally, we constructed novel in silico prediction systems, one of which can evaluate potential bitter molecules with high accuracy (AUC = 0.850) using consensus scoring based on the structure-based ML tools OnionNet, GNINA and BAPA.



