Estimation of uncertainty in Brain Tumor segmentation using modified multistage 3D-UNet on multimodal MRI images
DOI:
https://doi.org/10.62110/sciencein.jist.2024.v12.802Keywords:
Brain Tumor Segmentation, uncertainty, MRI Images, 3D UNet, Deep LearningAbstract
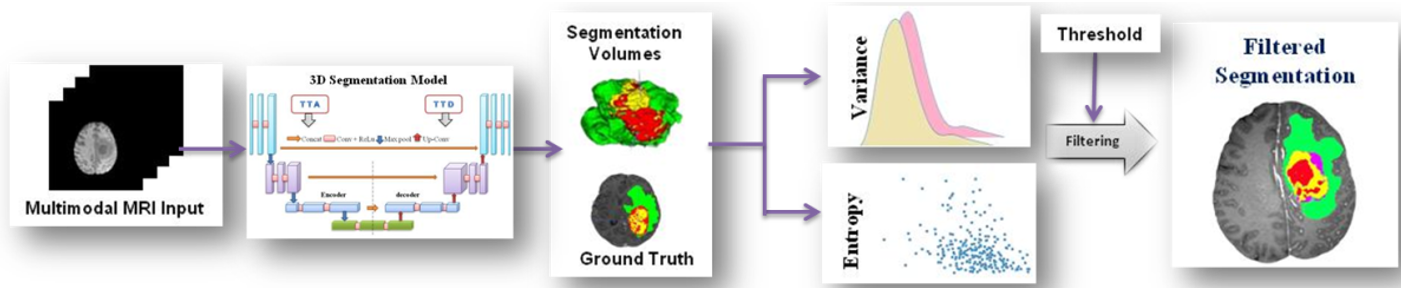
Automated brain tumor segmentation is challenging due to the tumor tissues' shape, size, and appearance. Various methods used multi-mode MRI scans to segment sub-regions of brain tumors. 3D CNN methods improved performance in recent years, but most methods do not use uncertainty information in segmentation. For reliability and understanding, model prediction is vital for clinical decisions. This work studies three models namely 3D-UNet, Modified 3D-UNet, and Modified Multistage-3D-UNet for brain tumor segmentation. MRI volume bias correction and normalization were carried out using z-score normalization. Two patch generation strategies reduce memory use and class imbalance. Voxel-wise uncertainty evaluation was made for aleatoric and epistemic uncertainties using test time augmentation and dropout, respectively. Variance and entropy are used to measure the uncertainty of the modified multistage-3D-Unet segmentation model from ground truth. Variance creates separate uncertainty maps for each tumor sub-regions, whereas, entropy provides only global information. Uncertainty is used to filter miss-segmented predictions and improve accuracy. Uncertainty awareness increases model accuracy with dice scores of 0.93, 0.91, and 0.83 for tumor sub-regions WT, TC, and ET respectively.
URN:NBN:sciencein.jist.2024.v12.802
Downloads
Downloads
Published
Issue
Section
URN
License
Copyright (c) 2024 Bhavesh Parmar, Mehul Parikh

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Rights and Permission







